This research was published in the INSPIRED: A Publication of the New Hampshire Agricultural Experiment Station (Fall 2024)
Researchers: A. Strickland, B. Y. Lee and B. L. Brown
Eastern oysters (Crassostrea virginica) are important to estuarine ecosystems in New England, providing habitat, buffering extreme weather, and sequestering nutrients such as N, P, and C. Oyster restoration supports local fisheries and aquaculture industries. However, in New Hampshire's Great Bay Estuary, oyster populations are at risk due to diseases like MSX and Dermo, caused by infectious agents Haplosporidium nelsoni and Perkinsus marinus, respectively. Improved assessment of the presence and distribution of these pathogens in GBE waters is key to strategic and effective oyster restoration efforts.
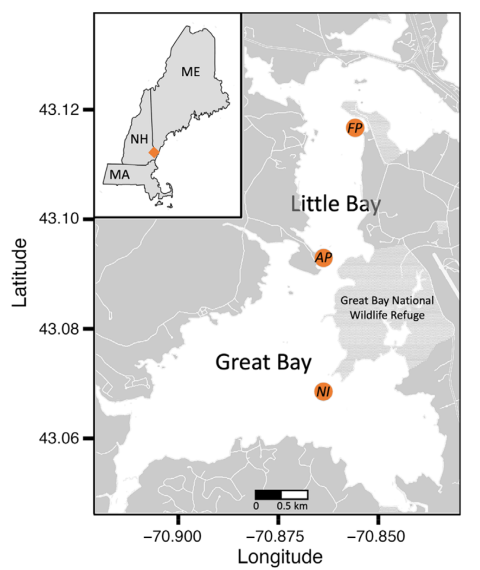
Fig. 1. Great Bay Estuary (orange diamond in regional inset of US New England states bordered in black). Sampling sites (orange points) included an oyster reef at Nannie Island (NI), an oyster farm near Fox Point (FP), and a site lacking a substantial oyster population between the other two sites near Adams Point (AP).
Background and Key Concepts
One challenge to the successful restoration of New Hampshire's declining wild oyster population is the presence of shellfish diseases. MSX and Dermo are protozoan pathogens affecting Eastern oysters. MSX is caused by the pathogen Haplosporidium nelsoni, and Dermo is caused by the pathogen Perkinsus marinus. Both diseases have contributed to declines in oyster populations in various estuaries, including Great Bay Estuary (GBE). Understanding the prevalence and transmission mechanisms of these pathogens is crucial for effective management and restoration efforts. This study used molecular techniques to detect and quantify pathogen DNA in GBE water samples.
Methodology
The study was conducted at three sites in (Fig. 1) GBE: a native oyster reef at Nannie Island, an oyster farm in Little Bay, and a reference site at Adams Point with no detectable oysters. Weekly water samples were collected from June to November 2020. DNA was extracted, and disease intensity was quantified using a quantitative-competitive PCR assay developed at UNH that allows for the simultaneous detection of Haplosporidium nelsoni and Perkinsus marinus DNAs. The chosen sites represented different oyster population scenarios, providing a comprehensive overview of pathogen presence in various contexts within GBE.
Key Findings
- Waters near Eastern oyster habitat in New Hampshire’s Great Bay estuary exhibited higher, more variable concentrations of DNA from the pathogens Haplosporidium nelsoni (which causes the disease MSX) and Perkinsus marinus (which causes the disease Dermo).
- Significant peaks for H. nelsoni DNA were observed from late July through August, while higher levels of P. marinus DNA were detected in June and maintained throughout the summer in real-time.
- Detection of H. nelsoni DNA in oyster larvae suggests potential pathways for disease transmission.
About the Co-author

Bonnie Brown, Professor of Biological Sciences
Contact information: Bonnie.Brown@unh.edu, FindScholars profile

Tonging for oyster samples at experimental reefs.
Discussion of Findings
The study found significant levels of pathogen DNA at sites with oysters, with notable seasonal peaks (Fig. 2). High concentrations of H. nelsoni DNA were observed in late July through August, primarily at the native reef site, suggesting that wild oysters might be more susceptible to MSX. The farm site, which housed MSX-resistant oyster strains, showed relatively lower levels of H. nelsoni DNA. This indicates that breeding and deploying resistant strains can mitigate the impact of MSX.
Perkinsus marinus DNA was consistently higher than MSX at sites with oysters compared to the reference site, with peak levels detected in June and maintained throughout the summer. The persistent high levels of P. marinus DNA highlight the need for ongoing monitoring and management of Dermo disease and for the development of a Dermo-tolerant strain.
The study also discovered H. nelsoni DNA in planktonic oyster larvae, suggesting that larvae could be potential vectors for disease transmission. This finding is significant as it underscores the importance of monitoring not only adult oysters but also larval stages to understand the full scope of pathogen dynamics.
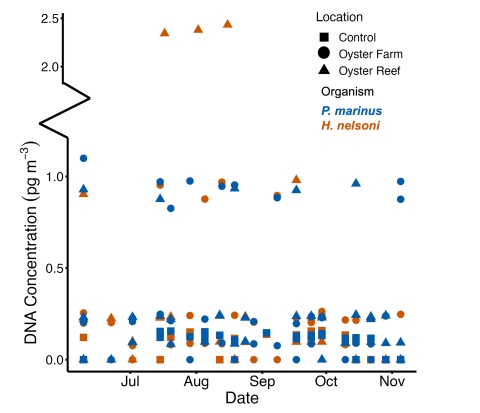
Fig. 2. Trends in water concentration of H. nelsoni and P. marinus DNA in Great Bay Estuary, New Hampshire.
Strategic Recommendations and Conclusion
Continuous monitoring of pathogen levels in estuarine waters is crucial for effective disease management. Restoration projects should consider incorporating pathogen-resistant oyster strains to mitigate disease impacts. Further research into the transmission mechanisms of MSX and Dermo, particularly the role of oyster larvae and other potential intermediate hosts, is essential for developing comprehensive restoration strategies.
The findings of this study underscore the importance of understanding pathogen dynamics in GBE for successful oyster restoration. High levels of MSX and Dermo pathogens in waters associated with oyster habitats highlight the need for integrated disease management approaches, including continuous monitoring, strategic use of resistant strains, and further research into transmission pathways. These integrative strategies are vital to support the recovery and sustainability of oyster populations in GBE. Ongoing research and adaptive management practices are likely to increase the likelihood of long-term health and viability of New Hampshire's estuarine ecosystems.
This material is based on work supported by the NH Agricultural Experiment Station through joint funding from the USDA National Institute of Food and Agriculture (under Hatch award number 1023564) and the state of New Hampshire.
