Signal transduction is the study of how cells control their own and each other's behaviors through physical (light, sound) or chemical (hormone, neurotransmitter) signals. Signal transduction research is an intensely active field of research. UNH faculty are engaged in various research programs that employ biochemical, molecular, cellular, and genetic/genomic approaches to understand cellular communication.
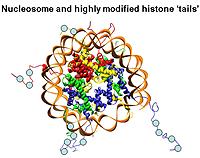
Feixia Chu — Epigenetics of histone post-translational modifications
Chromatin, the complex of genomic DNA and its associated proteins, constitutes the converging focus of various signaling pathways, and its structure defines the inheritable identity and fate of the cell. Histones are major chromatin proteins that are responsible for packaging genomic DNA into higher-order structures. Modifications of histone proteins can regulate effector protein binding or directly modulate chromatin structure, hence playing an important role in mediating various cellular responses to environmental influences. Using quantitative mass spectrometry, the Chu lab captures a panoramic view of the histone epigenetic “landscape,” and how it changes dynamically in response to cellular signaling processes such as differentiation and the DNA damage response. The functional significance of histone modifications during specific cellular processes is also independently approached from both biochemical and cell biological perspectives.
Feixia Chu
Rudman Hall, Room 306
Phone: (603) 862-2436
Email: feixia.chu@unh.edu

Rick Cote — Visual transduction pathway in retinal rod and cone photoreceptors
Rick Cote’s lab studies the visual signaling pathway in retinal rod and cone photoreceptors, focusing on the regulation of photoreceptor phosphodiesterase (PDE6) during light activation of the phototransduction cascade. The experimental tools we rely on include a variety of molecular, enzymological, and cell biological approaches, along with collaborations with the Thomas laboratory (structural genomics of PDEs), with the Chu laboratory (proteomic discovery of the PDE6 protein "interactome"), and with the Laue laboratory (biophysical approaches to PDE6 structure/regulation). Ultimately, our biomedical research will contribute to the development of therapeutic approaches to intervene in retinal diseases.
Rick Cote
Rudman Hall, Room 379
Phone: (603) 862-2458
Email: rick.cote@unh.edu
Lab website: www.cotelab.org

Estelle Hrabak — Signal transduction in Arabidopsis: regulation by post-translational modifications
Estelle Hrabak's lab uses genetic and biochemical approaches to study enzymes involved in signal transduction in plants. Our primary study organism is Arabidopsis thaliana. The two major projects in the Hrabak lab focus on protein phosphatases and palmitoyltransferases; both enzymes are encoded by large gene families in Arabidopsis. Protein phosphatase 2A (PP2A) is known to regulate many hormone response pathways in plants and likely has other functions. For example, using a reverse genetic approach, we found that some pp2a mutants have altered responses to sodium stress. Currently, we are dissecting the cellular and subcellular basis of this mutant phenotype. Palmitoyltransferases (PAT) are integral membrane proteins that acylate their substrates by covalent attachment of the fatty acid palmitate. Acylation can have major effects on the subcellular localization, half-life, or activity of the substrate proteins. We are interested in identifying the substrates of PATs, determining in which cellular membrane(s) each PAT resides, and characterizing the phenotypes of pat mutants. Our research's long-term goal is to thoroughly understand the basic biology of plants for application to biotechnology or agriculture.
Estelle Hrabak
Rudman Hall, Room 105
Phone: (603) 862-0716
Email: estelle.hrabak@unh.edu
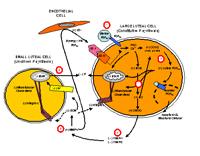
Paul Tsang — Female reproductive physiology
Paul Tsang's lab studies female reproductive physiology using the sheep, cow and fish animal models. The varieties of experimental tools we use include whole animal, cellular, molecular and cell culture approaches. In sheep, we study the molecular mediators of corpus luteum regression initiated by prostaglandin F2 alpha (see accompanying cell model). In the cow, we are profiling the molecular mediators associated with the angiogenic switch in the ovarian follicle and the corpus luteum. In sharks and skates, we study these elasmobranchs' life history, including reproduction, age and growth, and population structure. Our collaborators include John McCracken (sheep; University of Connecticut), Marsha Moses (cows; Children's Hospital Boston) and James Sulikowski (sharks and skates; University of New England). Ultimately, our research has applications in biomedicine (infertility and cancer) and fishery management.
Paul Tsang
Rudman Hall, Room 187
Phone: (603) 862-3479
Email: paul.tsang@unh.edu
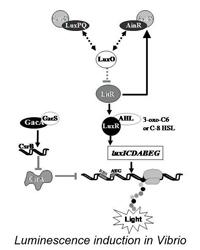
Cheryl Whistler — Transition of bacteria from the planktonic to the host lifestyle
The Whistler lab investigates hierarchical signal transduction systems that trigger the transition of certain bacteria from the planktonic to the host lifestyle. During host colonization, bacteria must coordinate several traits to overcome host defenses and establish an infection. To do this, they sense signals from their environment and then appropriately regulate gene expression. Several bacterial species utilize a classical two-component signal transduction system comprised of the GacS sensory kinase and the GacA response regulator. Interestingly, this signaling system can result in either disease or establishment of a beneficial symbiotic association. We are characterizing the transcriptional and translational regulatory mechanisms by which GacS/GacA governs bacteria's ability to activate genes by cell density in response to chemical pheromones (quorum sensing). Also, research is directed at understanding how changes in the host's temperature induces a transition to the pathogenic state. Our lab also focuses on how symbiosis and virulence are regulated by complex gene networks through the use of whole-genome transcriptomics and proteomics approaches. Our goal is to understand how bacteria integrate horizontally acquired colonization genes within these ancient regulatory networks to adapt to new environments.
Cheryl Whistler
Rudman Hall, Room 208
Phone: (603) 862-2359
Email: cheryl.whistler@unh.edu