No higher organism lives in isolation from a myriad of microbes. Associating microbes can either secure host nutrients by siege (causing disease) or by mutual consent (sometimes promoting the health of their partners). At the same time, both plant and animal hosts employ barriers to infection by pathogens, obstacles that challenge foe and friend alike and shape the adaptive evolution of microbial populations. The complexity of these different associations provides an exciting framework for discovery. A number of research programs at UNH explore this interface providing interdisciplinary training in microbiology, ecology, evolution, molecular biology, biochemistry, genetics, genomics and bioinformatics, with long-term applications of relevance to both applied and basic science.
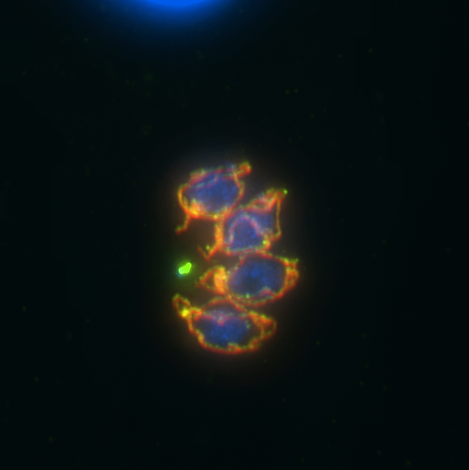
Vicki Jeffers – Epigenetic regulation in a protozoan parasite, Toxoplasma gondii
Toxoplasma gondii is a protozoan parasite that infects approximately 25% of people in the United States. The acute disease caused by this parasite is a major concern for immunocompromised individuals however, the chronic infection most commonly manifests as tissue cysts that reside in the brain of the infected individual. Toxoplasma is also a major veterinary concern, causing high rates of stillbirths in sheep and goats. There are currently no drugs available to treat the chronic stage of the infection and an infected person will carry the parasites in their brain for the rest of their life.
Epigenetic regulation of gene expression through the modification of histones is known to be critical for parasite viability, pathogenesis and the ability to switch between acute and chronic stages. The Jeffers lab is interested in the role of histone acetylation in this process, specifically the “reader” proteins that bind the acetylated histones and recruit other chromatin modifiers or transcriptional complexes. The bromodomain is one of these reader modules, and Toxoplasma has cache of these proteins, many of which are unique to this family of parasites, so their function is unknown. We use a wide variety of genomic, proteomic, molecular and biochemical techniques to discover the essential function of this group of proteins in Toxoplasma. Our ultimate goal is to identify small molecule inhibitors that specifically disrupt parasite bromodomain function to develop new, more effective anti-parasitic therapies.
Vicki Jeffers
Rudman Hall, Room 212
Phone: (603) 862-2103
Email: victoria.jeffers@unh.edu
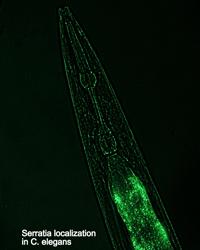
W. Kelley Thomas — Evolution of nematode-bacterial symbioses
The nematode Caenorhabditis elegans is a premier model organism in biology. In contrast to the tremendous scientific infrastructure built around the Caenorhabditis, our knowledge and understanding of the ecological role of this organism remains almost non-existent. The Thomas lab has recently discovered an association between a Caenorhabditis and a bacterium (Serratia) that forms a symbiotic relationship resulting in an insect-pathogenic complex. This complex is similar in function to entomopathogenic nematode-bacterial complexes that function as bio-control agents. Our work seeks to better understand how the nematode and bacterium communicate to function as a complex. This work benefits from the extensive knowledge and genomic toolbox available for both the Caenorhabditis and the bacterium.
W. Kelley Thomas
Gregg Hall, Room 448
Phone: (603) 862-2470
Email: kelley.thomas@unh.edu

Louis Tisa — Host-microbe interactions in symbiosis and pathogenesis
The Tisa lab is interested in genome-wide approaches toward studying host-microbe interactions in symbiosis and pathogenesis. These relationships have common mechanisms for establishment of the association including the ability to overcome the host defenses. The roles of microbial behavior, signal molecules, natural products, and signal transduction in host-microbe interactions are areas of focus. Current work centers on two major model systems: plant-microbe and microbe-nematode interactions. The plant-microbe system is centered on the Frankia-actinorhizal plants, which has an infection process to develop a root nodule structure; this work is aided by several international collaborations including laboratories in France, India, Tunisia, Egypt, Japan, and Argentina. Our lab also explores microbe-nematode interactions, concentrating on Photorhabdus-entomopathogenic nematodes (Heterorhabditis) association, which is a useful biological control agent for several insect pests. Nematode development and reproduction has an obligate requirement for their bacterial symbiont. The bacterium Photorhabdus maintains two distinct life styles as a nematode symbiont and as an insect pathogen. Thus, it provides a unique and beneficial model system for host-microbe interactions and for studying the genes controlling pathogenesis, symbiosis, and the switch between these two states. We are also interested in understanding the roles that bacterial motility, biofilm formation and signal transduction play in insect pathogenesis and nematode symbiosis. We also collaborate with the Thomas lab (UNH) on the newly identified microbe-nematode interaction (Serratia-C. briggsae) that forms an entomopathogenic complex.
Louis Tisa
Rudman Hall, Room 289
Phone: (603) 862-2442
Email: louis.tisa@unh.edu
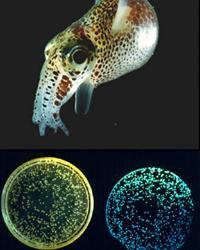
Cheryl Whistler — Squid-Vibrio mutualism and pathogenesis
Research in the Whistler lab seeks to elucidate the molecular mechanisms that allow host association by both pathogens and symbionts, using model organisms in the genus Vibrio. Our subjects of study are an elegant mutualistic symbiosis between a bioluminescent bacterium Vibrio fischeri and its squid host, and an environmentally transmitted pathogen Vibrio parahaemolyticus for which virulence is elusive and poorly defined. We apply a broad range of molecular genetic and genomic approaches in this work to identify novel colonization factors, define how microorganisms coordinate gene expression during the transition from a free-living to host associated lifestyle, and to better understand how microorganisms adapt and evolve to these associations. We are particularly interest in how horizontally acquired factors that facilitate host range expansion and adaptation are integrated into ancient regulatory networks that coordinate core colonization functions. Specific projects include the interrogation of global regulatory networks that coordinate transcription and translation of colonization factors on a whole genome basis, the development of alternative host models of infection for discovery of disease mechanisms, comparative genomics and transcriptomics between symbionts and pathogens, and the application of experimental evolution to investigate adaptive mechanisms of symbiosis (in collaboration with the Cooper lab). We believe a better understanding of the forces shape the outcome of associations will provide new avenues for enhancing health, and capitalizing upon microbial interactions for our benefit.
Cheryl Whistler
Rudman Hall, Room 208
Phone: (603) 862-2359
Email: cheryl.whistler@unh.edu