The enzymatic, scaffolding, trafficking and regulatory functions of proteins are carried out on the platform of their three-dimensional structures. Mechanistic insights on diverse cellular processes can be derived from the characterization of protein structures, including their 3D structure and hydrodynamics, spatial and temporal regulation of protein interactions, and protein glycosylation. A number of UNH research programs are addressing the structure/function relationship of proteins and protein assemblies with diverse spectroscopic approaches utilizing centrifugation, mass spectrometry, and chemical or fluorescent labeling.
Feixia Chu — Structural determination of protein assemblies
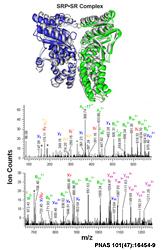
Protein assemblies play pivotal roles inside the cell, functioning as highly connected and dynamic modules to carry out cellular processes. Knowledge of the molecular recognition (or ‘molecular sociology’) among proteins within these assemblies is critical to understanding the orchestration of sophisticated cellular processes. The Chu lab is using chemical cross-linking in conjunction with mass spectrometric analysis to elucidate structural information on protein assemblies. Data from such biochemical analysis can be integrated through computational modeling to reveal protein interaction surfaces and novel conformations of protein complexes.
However, accurate identification of chemical cross-linked species has proved challenging. Towards this front in proteomics research, our lab is making new chemical probes, designing novel separation strategies, and developing new bioinformatic tools. In collaboration with multiple groups, we are using this approach to provide a mechanistic understanding of key complexes in visual transduction, transcriptional regulation, protein translocation and nuclear receptor signaling.
Feixia Chu
Rudman Hall, Room 306
Phone: (603) 862-2436
Email: feixia.chu@unh.edu
Rick Cote — Structural approaches to understanding the visual signaling pathway
Light stimulation causes the activation of the cGMP phosphodiesterase (PDE6), the central effector of the visual transduction pathway. Precise regulation of the lifetime of PDE6 activation is required to control the sensitivity, amplitude and kinetics of the light response. The Cote lab focuses on understanding the sequence of steps by which transducin relieves the inhibited state of the PDE6 holoenzyme, as well as identifying other proteins which may form a multi-protein complex with PDE6 to further regulate its hydrolytic activity.
Several putative PDE6 binding partners have been reported, but the significance of most of these interactions for the phototransduction mechanism is not known. In collaboration with the Chu lab (UNH), we are using proteomic tools to study the ‘interactome’ of PDE6 and to characterize the PDE6 signaling complex in its dark-adapted, transiently activated, and persistently activated states.
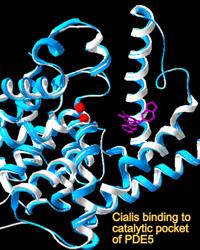
Another active area of investigation relates to the structural and functional relatedness of the photoreceptor PDE6 enzyme family with PDE5, prevalent in vascular smooth muscle and a therapeutic target of drugs increasingly used for treatments of the cardiovascular and urogenital systems. Adverse visual effects can accompany administration of PDE5 inhibitors due to the lack of selectivity of most drugs for PDE6. Whereas PDE5 is readily expressed as a recombinant protein, efforts to express the structurally and functionally related PDE6 have met with failure. Using replacement of evolutionarily conserved amino acids that differ between the PDE5 and PDE6 families, we are identifying the amino acids that confer drug specificity and catalytic efficiency of PDE6.
Rick Cote
Rudman Hall, Room 379
Phone: (603) 862-2458
Email: rick.cote@unh.edu